In the next series of blog posts we are going to look at Christian questions about evolution. In my last post we saw that two of the Christian views of creation, Young Earth Creationism and Old Earth Creationism, disagree with the scientific account of the mechanism of creation. Because of this we need to ask, is evolution true? Perhaps the strongest recent challenge to evolution has been the Intelligent Design movement. For the rest of this post we are going to look at Intelligent Design and ask if it is a better explanation than the scientific view of evolution by natural selection.
What is Intelligent Design? Intelligent Design supporters make three claims.
- Certain features of the universe and of living things are best explained by an intelligent cause, not an undirected process such as natural selection.
- Intelligent design is detectable based on empirical observations using objective criteria.
- Reference to intelligent design is legitimate within a scientific explanation of events in natural history.

How might we detect God’s supernatural intervention during the course of natural history? If God has intervened supernaturally during the course of natural history, one would expect to find gaps in the chain of natural explanations. And in fact there are gaps. Some examples are the origin of life, the origin of biological information, the origin of complex biological structures, the origin of the universe, and the fine-tuning of the laws of nature for life. So those gaps could represent places where God has intervened supernaturally. However, all scientific explanations are underdetermined by the data so it is not surprising to find gaps. Furthermore, science has a history of filling in the gaps. Therefore the presence of gaps alone doesn’t prove that the gaps are due to direct action by God.

So how do we decide between these two options? Enter a man named William Dembski. In his book, Intelligent Design: The Bridge Between Science and Theology (InterVarsity Press, 2002), he outlined a design filter to help decide whether gaps have a natural explanation or whether they are due to the actions of an intelligent designer. The filter is basically a decision tree. Beginning with some event in natural history the first decision is whether the event was a necessity or whether it was contingent. An event is necessary if it is required by natural law. For example, if I drop a pencil it will fall until it hits a solid object such as a table or the floor. The fall of the pencil is necessary because of Newton’s law of gravitation. On the other hand the decision to drop the pencil was a contingent event since I didn’t have to drop the pencil.
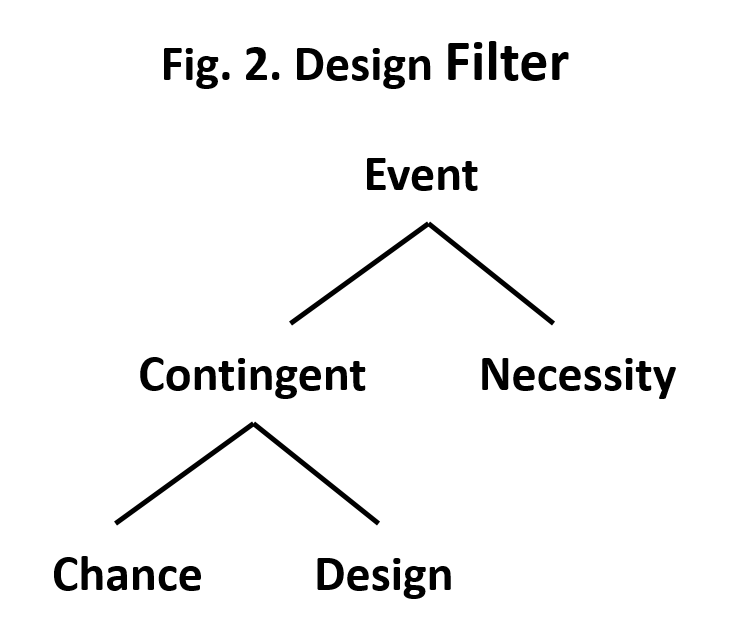
If an event is contingent the next choice is between chance and design. In my pencil example, the initial event of dropping the pencil could have resulted from my conscious choice (design) or it could have been an accident (chance). In Dembski’s filter the decision between choice and design is based on probability, with the choice of design requiring that the event has a very low probability of occurring by chance. The choice of design requires that the event have two features: complexity and specification. Consider a random selection of 8 letters: axlmqyrz. If we allow the same letter to be selected more than once, then there are 26 possibilities at each position in the sequence of letters or 209 billion combinations of the eight letters. So the first requirement of complexity is met but there is no specificity, in other words the sequence of letters have no meaning. We would then conclude that the set of letters were drawn by chance. On the other hand if we selected the following eight letters: decision. This sequence of letters spells a word and thus does have meaning or specification. The probability of this sequence of letters occurring by chance is very low (one in 208 billion). Based on his decision tree we would conclude that drawing letters spelling the word decision occurred by design rather than by chance. Dembski refers to this situation as specified complexity.

Mount Rushmore is another example of specified complexity. The faces of the presidents carved into the mountainside clearly exhibit design because they are complex and they also have meaning or specification. On the other hand, the surrounding mountainside displays complexity due to variations in composition and effects of erosion but there is not specification or meaning. Using Dembski’s filter we could then conclude that the faces of the presidents are there by design not by chance.
There are two types of intelligent design: biological and cosmological. Biological intelligent design asserts that some biological structures are too complex to have been produced by natural selection. Cosmological intelligent design asserts that science is not able to answer certain basic questions about the origin of the universe and its basic properties. In this post I’m going to focus on biological intelligent design because it represents a challenge to the dominant scientific paradigm of evolution by natural selection. I will talk about cosmological intelligent design in a later post.

Biological Intelligent Design. In 1996, Michael Behe, a Biochemistry professor at Lehigh University, published a book called Darwin’s Black Box. He argued that cells were full of complex molecular machines and that natural selection was not able to explain the origin of these machines. Behe defined a parameter called irreducible complexity as an important feature of these complex systems. An irreducibly complex system displays two main features:
- It is complex with many interacting parts.
- All of the parts are necessary for function. The removal of any one part causes the system to stop working.
Behe goes on to argue that an irreducibly complex system can’t be created by natural selection because all of the parts must be present before the system starts to function. Natural selection works through a stair step process in which each step represents a small improvement or a movement towards a new function that can be selected for. In the case of an irreducibly complex structure there seems to be no way to select for intermediate steps because they are non-functional. Therefore Behe argues that intelligent design is a better explanation than natural selection.
Behe discussed several examples of irreducibly complex systems including: the bacterial flagellum, the human blood clotting apparatus, the machinery for transporting materials within cells, the immune system, and the biochemical pathway for the synthesis of ATP the energy currency in cells. Behe argued that these biological machines were too complex to have been created through natural selection and thus required the intervention of an intelligent designer.
Human blood clotting apparatus. Let’s look at the human blood clotting apparatus as a well-characterized example of an irreducibly complex system. When you cut your finger it begins to bleed but after a while it stops. What makes it stop? The answer is that a protein in blood called fibrin aggregates and forms a clot that plugs the leak.
How does a clot form? There is a precursor form of fibrin in blood called fibrinogen. This protein accounts for about 3% of the protein in the liquid part of blood. In response to injury fibrinogen is converted to fibrin through the action of a protease called thrombin. A protease is an enzyme which acts like a kind of scissors to clip off pieces of a protein molecule. In this case the pieces that are clipped off mask a sticky part of the fibrin molecule that allows individual fibrin molecules to stick together and form the blood clot.
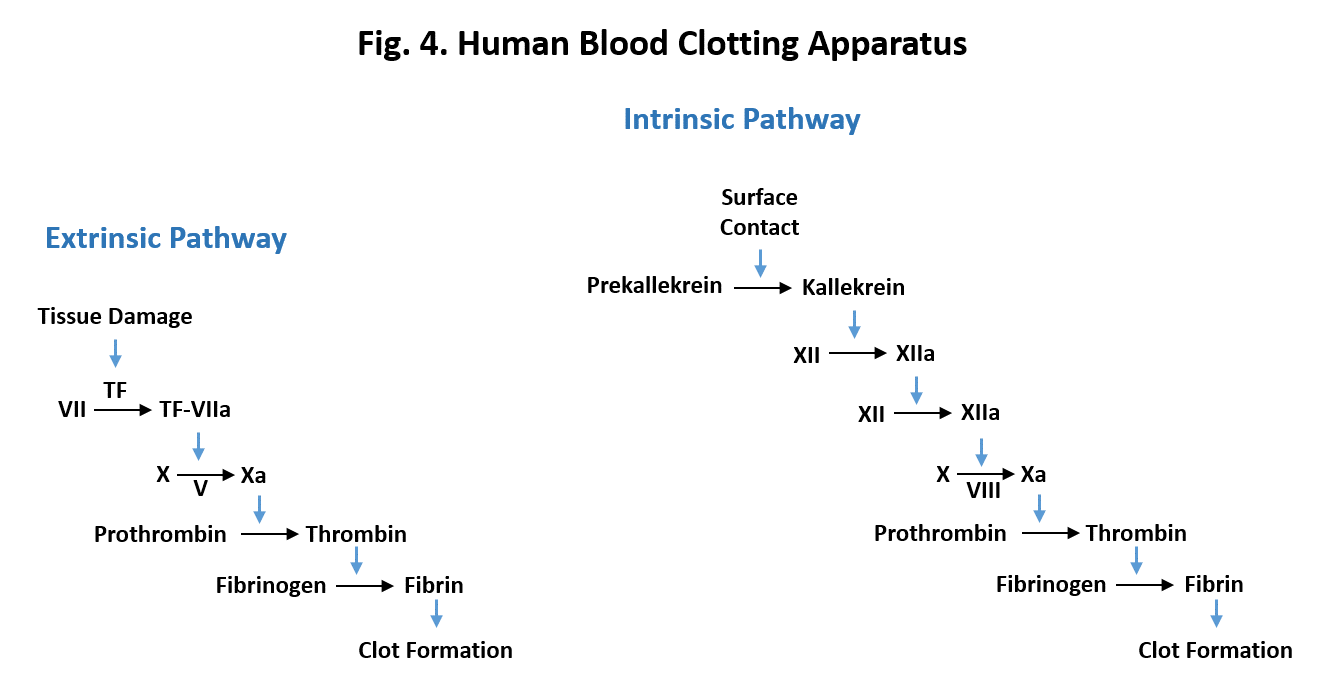
How does clot formation get started? There are two ways called the extrinsic and the intrinsic pathways. Both involve the conversion of prothrombin which is inactive as a protease to active thrombin. The extrinsic pathway (Fig. 5, left) begins when injury to tissue exposes a protein on the surface of cells called Tissue Factor (TF). This sets off a chain of events in which Tissue Factor combines with and activates Factor VII which in turn activates Factor X which then converts prothrombin to thrombin. Factors VII and X are both proteases which activate their targets by clipping off parts of the inactive molecule. In the diagram, the letter “a” after the factor indicates the active form. Factor V is a protein helper in the activation of Factor X.
The intrinsic pathway (Fig. 5, right) also sets off a chain reaction but there are more components in the pathway. The pathway is initiated when blood comes in contact with a foreign surface due to injury. This results in the activation of a protease called Kallikrien which sets off the chain of events. Factors XII, XI, and IX are proteases whereas factor VIII is protein that acts as a helper in the activation of Factor X.
Why are the two pathways complex with so many steps? Let’s first consider the extrinsic pathway. The major components in the pathway (Factor VII, Factor X and Thrombin) are all enzymes. Because of this one Factor VII molecule is able to activate many molecules of Factor X. Once activated Factor X is able to convert many molecules of prothrombin to thrombin which then converts many fibrinogen molecules to fibrin which then aggregates to form the clot. This creates a snowball effect in which a small beginning leads to a large ending. So the purpose of the cascade of events in the extrinsic pathway is to amplify the signal from the tissue damage and to speed up the response to the injury. Similarly the multiple steps in the intrinsic pathway also amplify the response to contact activation. Amplification of the response to injury is important for the human circulatory system because it is under high pressure and thus a lead needs to be plugged quickly to prevent excess bleeding.
Both of the blood clotting pathways meets Behe’s criteria for an irreducibly complex system.
- Each pathway has multiple interacting parts that participate in the blood clotting process
- Each pathway fails with the removal of any one clotting factor. As an example, deficiencies in Factors VIII and X in the intrinsic pathway are responsible for the two most common types of Hemophilia.
Could the human blood clotting system have arisen through evolution by natural selection? Russell Doolittle has devoted his career to studying the evolution of the blood clotting cascade in humans and other vertebrate animals (animals that have a backbone). He has studied the appearance of clotting factors during the course of vertebrate evolution. His work has been greatly aided by the development of DNA sequencing technologies that have provided detailed information about the genes present in modern members of fish, amphibians, reptiles and mammals. The complete extrinsic clotting system is present in fish but the intrinsic clotting system is absent. During the transitions from fish to amphibians to reptiles to mammals there is a gradual appearance of the clotting factors needed for the intrinsic pathway. The intrinsic pathway is complete in mammals but not in amphibians and reptiles. During the intermediate stages of evolution (amphibians and reptiles) the clotting factors of the intrinsic pathway are used to enhance the function of the extrinsic pathway.
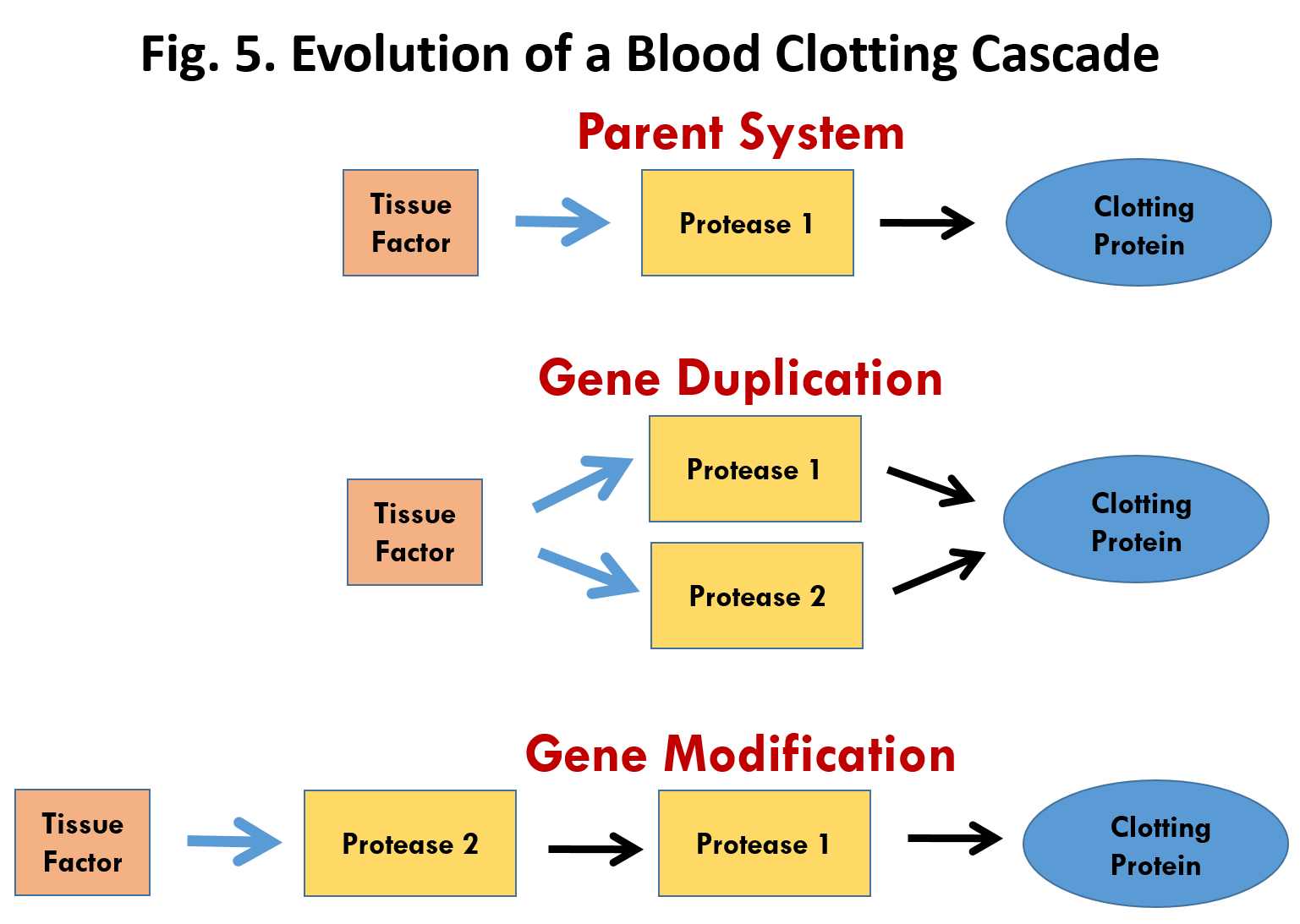
Doolittle’s work provides solid evidence for the evolution of the intrinsic pathway by natural selection. But what about the origin and evolution of the extrinsic pathway. We don’t yet know how the pathway developed during the transition from non-vertebrates to vertebrate animals. But here is a plausible mechanism. The large scale sequencing of genomes has revealed that most genes and the proteins that they code for can be grouped into families that are related in structure. These families have arisen through a well-known process called gene duplication. Gene duplication occurs occasionally during cell division when part of the DNA in a chromosome gets copied twice. Once gene duplication has occurred, the original gene continues with its normal function but the duplicate gene is free to undergo mutations.
How might gene duplication help to explain the origin of the Extrinsic Pathway? The first thing to notice is that all of the components in the pathway are proteases (protein-cutting enzymes) with the exception of Tissue Factor and Factor VIII. In fact they belong to the serine-protease gene family. Figure 6 illustrates a plausible scenario for the evolution of this pathway through gene duplication. The parent system consists of just three components (Fig. 6, top), a Tissue Factor, Protease 1 and a Clotting Protein. Tissue factor would be exposed upon injury and then bind to and activate Protease 1 which in turn would activate the Clotting Protein leading to aggregation and clotting. Duplication of the protease 1 gene would lead first to an intermediate system (Fig. 6, middle) with two identical proteases acting on the Clotting Protein. Both would be activated by Tissue Factor. Modification of the active site of Protease 2 by mutation could lead to a new arrangement (Fig. 6, bottom) in which the specificity of Protease 2 was changed allowing it to act on Protease 1 rather than the Clotting Protein. This new arrangement would amplify and speed up the clotting process and thus provide a basis for natural selection.
The scenario outlined above represents a plausible, although not proven, scenario for the evolution of the extrinsic blood clotting pathway beginning with a simple three component system. The process of gene duplication followed by gene modification could be repeated to add additional proteases thus further accelerating the response and providing a basis for natural selection at each step.
A scenario similar to that in Fig. 5 would also explain the evolution of the clotting factors in the intrinsic pathway. Again most of the components of the pathway belong to the serine protease family and thus could have arisen through gene duplication and subsequent modification as illustrated in Fig. 5. The only exception is Factor VIII which is closely related to Factor V (extrinsic pathway) and presumably arose through duplication of Factor VIII.
Conclusions.
- Irreducible complexity does not always rule out evolution by natural selection. We’ve seen that the blood clotting system meets Behe’s definition of an irreducibly complex system and yet there is good evidence that it arose through evolution by natural selection.
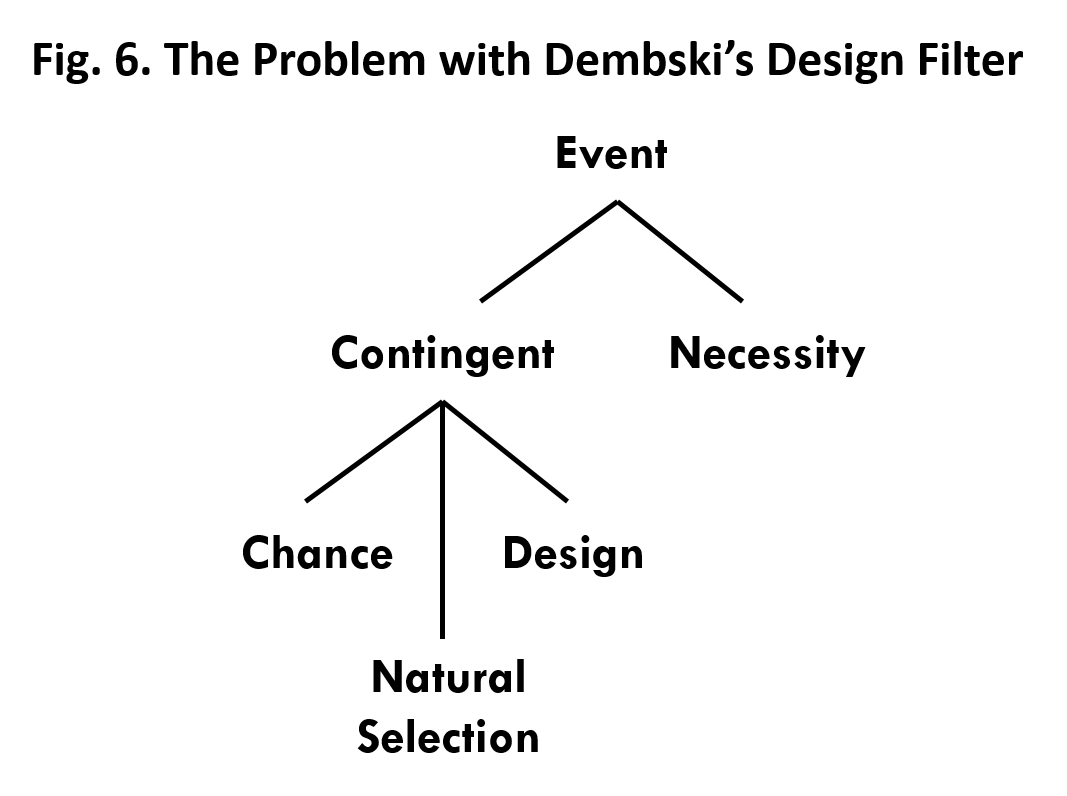
- Dembski’s design filter works through the process of elimination. It considers only two possible explanations for a contingent event, chance and design. If chance is eliminated then design is inferred because it is the only choice left. But, what we’ve seen is that natural selection is another viable alternative for the origin of at least some irreducibly complex biological systems. Natural selection gives the appearance of design without actually being design. The filter doesn’t provide a way of distinguishing between natural selection and design as the explanation for the origin of irreducibly complex biological systems.
- At the beginning of this post, I said that there were two explanations for gaps in the chain of natural explanations of events in natural history. They could be places where God has intervened supernaturally or they could be places where the natural explanation has yet to be discovered. What I’ve shown in this post is that neither Dembski’s design filter nor Behe’s notion of irreducible complexity produces a convincing argument for the supernatural action of God in these gaps.
- As a Christian I deeply believe that God is the Designer and Creator of life and the universe! But it is difficult to prove this using science alone. In a later blog post, I’m going to suggest a different approach to seeing God as the designer that combines God’s revelations in scripture and in nature (i.e. science).
Looking ahead: In my next post (March 19), we are going to look at the question: Does science in general and evolution in particular rule out God? On April 2 we will explore: Is evolution compatible with belief in God as the Creator?
Questions for further reflection:
- What is your view of the Intelligent Design argument? Is it more convincing than evolution by natural selection?
- Should Intelligent Design be taught in public schools as an alternative to evolution by natural selection?
- The Intelligent Design movement is neutral about who the designer is, but Christians would see God as the designer. Is it important to prove that God is the Creator through science alone?
Suggestions for further reading:
- Intelligent Design: The Bridge Between Science and Theology, William Dembski. InterVarsity Press, 2002.
- Darwin’s Black Box: The Biochemical Challenge to Evolution, Michael J. Behe. Free Press, 1998.
- Finding Darwin’s God: A Scientist’s Search for Common Ground between God and Evolution, Kenneth R. Miller. Harper Collins, 2007.
- Much of the work on Intelligent Design is being supported by the Discovery Institute in their Center for Science and Culture. There you can access more information about the design argument.
- Added by the editor: PDF with the titles and links to the posts in Tom Ingebritsen‘s Christianity and Science series. Yes, this was created to meet the requests of readers and will be updated. Your interest in and encouragement of this series is much appreciated.
Note: Originally titled Is Evolution True? The Case of Intelligent Design. Figure 4 Updated. Changes made on 3/14/2014 (5:04 pm).
Tom Ingebritsen is an Associate Professor Emeritus in the Dept. of Genetics, Development and Cell Biology at Iowa State University. Since retiring from Iowa State in 2010 he has served as a Campus Staff Member with InterVarsity Graduate & Faculty Ministry at Iowa State University

In your opening paragraph you say
“two of the Christian views of creation, Young Earth Creationism and Old Earth Creationism, disagree with the scientific account of the mechanism of creation. Because of this we need to ask, is evolution true? ”
Surely a better (or less biased) question would be “For which of these two accounts of the beginning of the universe (Creationism and the Scientific account) is there more reliable evidence?”
http://creation.com/scientific-evidence-for-creation
Thanks was interesting- towards the end though it feels like you’ve stopped questioning a bit. I get that you’re saying mutation but it’s oversimplifying – I.e it has to first exist to then duplicate etc etc.
Also you seem to draw a line between chance and accidental duplication being ‘selection’…
I like how you illustrated natural selection as a third option in Dembski’s filter. I think it would be helpful to expound upon the difference between randomness and selection since this is a major hangup for creationists (forgive me if you already did since I don’t recall what was previously posted). Natural selection appears different than chance because just like design it is nonrandom. Mutations might be chance events, but natural selection means that beneficial mutations have an increased probability of being fixed while harmful ones have a decreased probability. Taking the example of a random string of letters, natural selection is akin to rearranging them until a word is formed (though this is an oversimplification). Each given arrangement is random, but the output is not, because selection is nonrandom.
Christians are still often troubled by mutations being governed by chance events. They don’t see room for God if something is chance. However, chance only means that it is not predictable by natural laws. There are plenty of chance occurrences we encounter on a daily basis (the flip of a coin, the lottery, etc). If Creationists, at least of the Calvinist persuasion, can still treat these events as chance events while simultaneously seeing God’s sovereignty in determining their outcome, why don’t they apply the same logic to the chance occurrences that form the mutations that drive evolution?
Tim-
Thanks for your comments about the difference between randomness and selection. I agree with what you are saying. I also like your definition of chance as something that is not predictable by natural laws. The existence of chance in natural selection actually makes room for God’s providential action rather than a totally deterministic system. I’m going to address the issue of whether evolution is compatible with belief in God as the Creator in an upcoming blog (scheduled for April 2).
For those who might want to dig a little deeper into the question of the origin of the human blood clotting systems, here is a link to an expanded argument for the evolution of the cascade from Ken Miller (http://www.millerandlevine.com/km/evol/DI/clot/Clotting.html). You can see Michael Behe’s rebuttal to criticisms of his argument on the blood clotting systems at http://www.discovery.org/a/442. It is entitled “In Defense of the Irreducibility of the Blood Clotting Cascade”.
Thanks Dr. Ingebritsen for the comments on intelligent design. I think it’s worth pointing out that Michael Behe never argued that all elements of the blood clotting cascade form an irreducibly complex system. In fact, he never argued that the extrinisic or intrinsic initiation pathways of the cascade are entirely part of the irreducibly complex core of the system. Rather he defined the “system” that is irreducibly complex as the components after the “fork” in the pathway. As he wrote in Darwin’s Black Box:
“Leaving aside the system before the fork in the pathway, where some details are less well known, the blood-clotting system fits the definition of irreducible complexity. The components of the system (beyond the fork in the pathway) are fibrinogen, prothrombin, Stuart factor, and proaccelerin. in the absence of any one of the components, blood does not clot, and the system fails.” (Behe, Darwin’s Black Box, p. 86.)
In that regard, Dr. Doolittle’s comments about organisms that lack some components of the intrinsic pathway might be interesting, but they don’t address Behe’s argument, because they occur before the fork. For a summary of these problems, please see:
http://www.discovery.org/a/8561
http://www.discovery.org/a/14081
I commend you for offering an intriguing hypothesis about the evolution of the extrinsic pathway. Let’s take a look at it.
First, your hypothesis starts with 3 pre-existing functional: the tissue factor, a protease, and the clotting protein. This suggests the blood clotting requires an irreducibly complex core, which your hypothesis assumes, rather than accounts for.
But could this system work? Many of the intermediate steps in the blood clotting cascade are crucial for regulating the cascade, helping to ensure that clotting doesn’t run out of control (which could kill the organism), or fail to happen at all, and that clotting proceeds at the right pace. It’s nice to assume that a simpler system can work, but given that many of the intermediate steps of the cascade are important, I’m not sure if it’s plausible to simply assert the simpler system would work. You note that your hypothesis:
“represents a plausible, although not proven, scenario for the evolution of the extrinsic blood clotting pathway beginning with a simple three component system”.
It’s very fair of you that you acknowledge that your hypothesis is not proven, but I’m not sure if it’s fair to say that your hypothesis is “plausible,” because I’m not aware of such a simple system being demonstrated in the real world.
Second, your hypothesis then proposes mechanisms for adding proteases to an irreducibly complex system, but you don’t identify which protease in your hypothesis pertain to which parts of the real blood clotting cascade, so it’s hard to know if your hypothesis pertains to any of the components that Behe claims are irreducibly complex. But let’s assume for the sake of argument that your hypothesis does attempt address the origin of components that Behe claims are part of the irreducibly complex core of the cascade, “after the fork,” as he put it.
Behe often has quoted Darwin saying that features which cannot be built through “numerous, successive, slight modifications” are inaccessible by the Darwinian mechanism because they would require multiple parts to be present before giving any advantage. He calls these irreducibly complex structures. Jerry Coyne concurs: “It is indeed true that natural selection cannot build any feature in which intermediate steps do not confer a net benefit on the organism.” (Jerry Coyne, “The Great Mutator,” The New Republic (June 14, 2007).)
Now the classic rejoinder is that one can evolve irreducibly complex features through exaptation (e.g., co-option of parts) rather than stepwise adaptation. Here again, however, research is suggesting that co-option of parts at the most basic levels—i.e., converting one protein into the function of a very closely related protein—could not occur given known probabilistic resources.
As you know, the basic model goes like this: some gene duplicates so one copy can perform the original function, and the duplicate copy can be “co-opted” to perform some new function. Perhaps these new duplicated genes can acquire new functions and be squeezed into networks or systems, thereby building irreducible complexity. This model is typically just asserted, as if it works, and is demonstrated through mere homology. But ID proponents have been willing to test it.
The duplication/divergence model of gene evolution ignores the problem of multimutation features which require multiple coordinated mutations to arise. There’s no question that genes can duplicate. The question is whether duplicated genes can traverse likely evolutionary pathways to acquire new functions before they are subfunctionalized. This is typically held to be a neutral walk—i.e., strictly random mutation—detached from selection pressure. However, population genetics imposes limits on how many specific neutral mutations a gene is likely to accumulate.
For starters, protein scientist Douglas Axe has published mutational sensitivity tests on enzymes in Journal of Molecular Biology[1] and found that functional protein sequences may be as rare as 1 in 10^77. That extreme rarity makes it highly unlikely that chance mutations alone could find the rare amino acid sequences that yield functional proteins:
See: http://www.evolutionnews.org/axediagram.jpg
According to Axe’s research, most enzymes sit at Point A, high atop their fitness landscape, and many amino acids must be present all at once to get high enzyme functionality. This suggests that the random walks required by co-option models of protein evolution would be very unlikely to find new adaptive peaks—i.e., to generate new protein folds.
But just how unlikely is it?
In 2010, Axe investigated this question, and published evidence indicating that despite high mutation rates and generous assumptions favoring a Darwinian process, molecular adaptations requiring more than six mutations before yielding any advantage would be extremely unlikely to arise in the history of the Earth.[2]
The following year, Axe published research with developmental biologist Ann Gauger describing the results of their experiments seeking to convert one bacterial enzyme into another closely related enzyme–one in the same gene family! That is the kind of conversion that evolutionists claim can easily happen. For this case they found that the conversion would require a minimum of at least seven simultaneous changes[3], exceeding the six-mutation-limit that Axe had previously established as a boundary of what Darwinian evolution is likely to accomplish in bacteria. Because this conversion is thought to be relatively simple, it suggests that the co-option model of evolution by gene duplication could not produce even many modest gene conversions.
In other experiments led by Gauger and biologist Ralph Seelke of the University of Wisconsin, Superior, their research team broke a gene in the bacterium E. coli required for synthesizing the amino acid tryptophan. When the bacteria’s genome was broken in just one place, random mutations were capable of “fixing” the gene. But even when only two mutations were required to restore function, Darwinian evolution got stuck, apparently unable to restore full function.[4] This is because it was more advantageous to delete a gene with low functionality or none than it was to continue to express it. This suggests that it is highly unlikely that the standard gene duplication model would produce new complex functions because gene duplicates are likely to be deleted before evolving some new function.
Other research corroborates these findings. Biochemist Michael Behe and physicist David Snoke have performed computer simulations and theoretical calculations showing that the Darwinian evolution of a functional bond between two proteins would be highly unlikely to occur in populations of multicellular organisms under reasonable evolutionary timescales. They found:
“The fact that very large population sizes – 10^9 or greater — are required to build even a minimal MR feature requiring two nucleotide alterations within 10^8 generations by the processes described in our model, and that enormous population sizes are required for more complex features or shorter times, seems to indicate that the mechanism of gene duplication and point mutation alone would be ineffective, at least for multicellular diploid species, because few multicellular species reach the required population sizes.”[5]
In other words, in multicellular species, Darwinian evolution would be unlikely to produce features requiring more than just two simultaneous mutations on any reasonable timescale or population size.
In 2008, Behe’s critics sought to refute him in the journal Genetics with a paper titled “Waiting for Two Mutations: With Applications to Regulatory Sequence Evolution and the Limits of Darwinian Evolution.” But Durrett and Schmidt found that to obtain only two specific mutations via Darwinian evolution “for humans with a much smaller effective population size, this type of change would take > 100 million years.” The critics admitted this was “very unlikely to occur on a reasonable timescale.”[6]
So basically, the research is showing that unguided evolutionary mechanisms—whether by Darwinian evolution (e.g., mutation and selection) or exaptation (e.g., duplication leading to co-option via neutral/random walk)—are very unlikely to produce new proteins, calling into question the gene-duplication models used by Doolittle and others.
Thanks for reading.
Sincerely,
Casey
References Cited:
[1] Douglas Axe, “Estimating the Prevalence of Protein Sequences Adopting Functional Enzyme Folds,” Journal of Molecular Biology, Vol. 341: 1295-1315 (2004); Douglas Axe, “Extreme Functional Sensitivity to Conservative Amino Acid Changes on Enzyme Exteriors,” Journal of Molecular Biology, Vol. 301: 585-595 (2000).
[2.] Douglas Axe, “The Limits of Complex Adaptation: An Analysis Based on a Simple Model of Structured Bacterial Populations,” BIO-Complexity, Vol. 2010(4):1-10.
[3.] Ann Gauger and Douglas Axe, “The Evolutionary Accessibility of New Enzyme Functions: A Case Study from the Biotin Pathway,” BIO-Complexity, Vol. 2011 (1): 1-17.
[4.] Ann Gauger, Stephanie Ebnet, Pamela F. Fahey, and Ralph Seelke, “Reductive Evolution Can Prevent Populations from Taking Simple Adaptive Paths to High Fitness,” BIO-Complexity, Vol. 2010 (2): 1-9.
[5.] Michael J. Behe and David W. Snoke, “Simulating Evolution by Gene Duplication of Protein Features That Require Multiple Amino Acid Residues,” Protein Science, Vol. 13: 2651-2664 (2004).
[6.] Rick Durrett and Deena Schmidt, “Waiting for Two Mutations: With Applications to Regulatory Sequence Evolution and the Limits of Darwinian Evolution,” Genetics, Vol. 180: 1501-1509 (November 2008).
Casey-
Thanks for your thoughtful response to my blog. In the blog, I presented a plausible scenario for the origin of the protease cascade of the intrinsic system. The scenario is not my idea but came from Ken Miller in his expanded argument for the evolution of the cascade (http://www.millerandlevine.com/km/evol/DI/clot/Clotting.html). You rightly point out that the system starts with three pre-existing protein components (tissue factor, protease 1 and a clotting factor, fibrinogen). So it’s reasonable to ask where these components came from. Here is what we know. As I said in the blog, the complete extrinsic clotting system is present in vertebrates (fish, amphibians, reptiles, birds and mammals). However, the nearest non-vertebrate relatives, the protochordates, do not form fibrin clots. They lack tissue factor, fibrinogen as well as the proteases (factor X and thrombin) necessary for the activation of fibrinogen.
Tissue factor is a membrane protein which not only activates Factor X but also localizes the clotting process to the site of injury. Tissue factor is a member of a family of membrane proteins called Class-2 cytokine receptors. While tissue factor is absent in protochordates, one or two other members of the family are present. Additionally, the transition from protochordates to fish (the earliest vertebrates) was associated with a tremendous expansion of the Class-2 cytokine receptor family.
While protochordates lack fibrinogen, they do have a fibrinogen-related protein that has a very similar overall structure to fibrinogen but lacks the regions necessary for activation of fibrinogen and the aggregation of fibrin. Protochordates also have members of the serine-protease family although they lack the specific proteases of the extrinsic blood clotting pathway.
All of this is to say that one can envision a pathway for the origin of a simple three-component precursor to both the extrinsic and intrinsic blood clotting pathways through gene duplication and subsequent modification. In the rest of the blog you raise questions about the viability of the gene duplication/modification model. But the argument you are making is basically an argument from elimination as I discussed in the blog. It is very difficult to prove a negative argument!
Dear Tom,
Thanks for your kind reply. I’m afraid that I really am not satisfied that you have answered my argument, however.
The fact that there are proteins in protochordates that are homologous to components of the blood clotting cascade in vertebrates does not therefore demonstrate an evolutionary pathway. What you, Ken Miller, and Rusell Doolittle do, repeatedly, is mix-up evidence for common ancestry as evidence for a viable evolutionary pathway. As Michael Behe observes:
“modern Darwinists point to evidence of common descent and erroneously assume it to be evidence of the power of random mutation” (Behe, The Edge of Evolution, p. 95).
Behe puts it even more clearly in Darwin’s Black Box:
“Although useful for determining lines of descent …comparing sequences cannot show how a complex biochemical system achieved its function — the question that most concerns us in this book. By way of analogy, the instruction manuals for two different models of computer put out by the same company might have many identical words, sentences, and even paragraphs, suggesting a common ancestry (perhaps the same author wrote both manuals), but comparing the sequences of letters in the instruction manuals will never tell us if a computer can be produced step-by-step starting from a typewriter….Like the sequence analysts, I believe the evidence strongly supports common descent. But the root question remains unanswered: What has caused complex systems to form?” (Darwin’s Black Box, pp. 175-176)
Thus, observations about comparative similarities in orthologous DNA sequences in no way refute irreducible complexity nor do they demonstrate a viable Darwinian evolutionary pathway. (Incidentally, when I was a graduate student at UC San Diego, I took a graduate seminar where Miller’s authority, Russell Doolittle, was brought in to tell us what was wrong with Michael Behe’s arguments for irreducible complexity in blood clotting. The main point of Doolittle’s lecture was to draw trees tracing protein relationships, all based upon mere comparison of sequence similarity. His argument that trees based upon sequence similarity somehow demonstrate Darwinian evolution and refute irreducible complexity left me unimpressed–it committed the very fallacy Behe warned about in Darwin’s Black Box. I came to a deeper realization at that point of just how strong Behe’s argument was.)
I just picked up a copy of 2013 Doolittle’s book “The Evolution of Vertebrate Blood Clotting,” and I have just had a little time to skim it. But it seems that in the dozen or so years since Doolittle taught my class, he’s still relying primarily upon sequence homology to make his case. From skimming the book, I do not see where Doolittle gives any kind of a stepwise evolutionary pathway to explain how one protein changed into another. (I am happy to be shown wrong; and if I am wrong, I’d love to know where Doolittle provides such an stepwise explanation.) Instead, Dr. Doolittle cites protein homology and then asserts that by standard gene duplication and subsequent mutation, the entire pathway could have evolved. Very little has changed.
You reply to my critiques of the “gene duplication + muttaion” model by saying: “It is very difficult to prove a negative argument!” If that is your position, then it would seem that the model of gene evolution by duplication (and subsequent mutation) is not a scientific hypothesis because it isn’t subject to falsification. Moreover, such a rebuttal does not address the empirically-based counterevidence to that model which I have presented.
The assumption behind the arguments that you, Miller, and Doolittle make is that if there’s some non-trivial level of sequence homology then it’s easy for mutation (and whenever possible, selection) to evolve the genes from a common ancestral genes. Is that a plausible assumption? Is it even possible to scientifically test a model which claims that similar genes evolved by mutation and selection from common ancestors?
Despite your suggestion to the contrary, I believe the model is testable. But it’s not a sufficient test of the model to merely cite protein homology, because that’s not evidence for a viable Darwinian pathway. Rather, a sufficient test of the model is to take highly similar genes with high sequence homology and ask whether an evolutionary conversion can be accomplished through an mutational pathway that does not require neutral “jumps” in complexity that are beyond what can be accomplished given available probabilistic resources (e.g., mutation rate, population size, generation time, time available from the fossil record, etc.). The papers I cited in my prior response (Axe, 2010, and Axe & Gauger, 2011), do just that.
I realize this is an ongoing debate, and that more research is necessary to fully answer these questions. But until we start asking the right questions, and stop adopting intentionally low standards of proof for demonstrating the evolution of complex systems, I fear these questions will not go away.
Thanks again for your time and thanks for reading.
sincerely,
Casey