In the previous post, Three Chromosomes in a Trench Coat, we learned that 1) a phenotype is the observable characteristics of an organism, 2) the phenotype is the result of interactions between the genome and the environment, and 3) the phenotype is not just the appearance of the organism but also includes where the organism is able to live. I also discussed why I think understanding about phenotypes is interesting and important. In this installment we will take a look at some of the nitty-gritty mechanics involved in turning a genotype into a phenotype. We will also learn something about epigenetics, a collection of processes that are integral to the realization of a phenotype from a genotype and a term that makes a semi-regular appearance the news in discussions about identical twins and cancer.
So just how does a genome become a phenotype? In science speak, an organism’s phenotype is the result of the expression of an organism’s genetic code and the influence of the environment. What exactly does this mean? How does a genetic code express itself when it doesn’t speak or do interpretive dance?

You made all the delicate, inner parts of my body and knit me together in my mother’s womb. (Psalm 139:13-16, NLT)
Let’s start with the Central Dogma (a.k.a. the basics of turning a gene into a functional product). The genetic code is a collection of instructions for proteins that are either building blocks or cogs in the biological machine. A gene is said to be expressed when the protein that it codes for is produced. First, the gene you want to make is transcribed by a complex of proteins called a polymerase that sits down on the DNA at the starting point of the gene of interest and makes a copy of the gene sequence using RNA. The transcript that is made is called messenger RNA or mRNA and has been in the news a lot during the last year because it is the basis for the vaccines against COVID-19 made by Pfizer and Moderna. The second step of gene expression includes moving the RNA to a different part of the cell, away from the chromosomes, where it binds to a complex of proteins and ribosomal RNAs (a.k.a. a ribosome) and is translated, or turned into a string of amino acids depending on the mRNA sequence. Interactions between individual amino acids cause the string to fold into a functional protein and it is transported to the location where it can do its predetermined job in the cell. The whole process is actually much more complex than I have described here. Anyone who wants more information about biological topics I’ve discussed in this post can use this very nice video glossary created by The National Human Genome Institute.
So how does this relate to our discussion of phenotype? The phenotype is created by expressing the appropriate genes at the appropriate time and place through development — the gradual process of increasing complexity from a single fertilized egg to a complete animal or plant. In the old days, circa 1650, this process was called “epigenesis” (Deichmann, 2016). Then, in 1942 Conrad Waddington coined the term “epigenetics” to refer to total sum of processes that lead to the production from the genotype to the phenotype. In other words, epigenetics are mechanisms that regulate gene expression during development. Epigenetic processes also have a role regulating gene expression in response to changes in the environment (something we will learn about in the next installment of this series). In current studies, epigenetic processes are defined as processes that can change the phenotype of an organism through multiple generations without changing the DNA sequence.
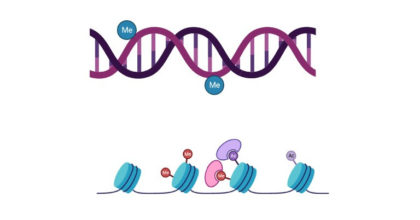
So, how do epigenetic processes work? There are 3 broad categories of epigenetic processes — DNA methylation, histone modification, and small noncoding RNAs. In a nutshell, DNA methylation and histone modifications generally inhibit gene expression by blocking transcription and preventing mRNA from being made. In the case of DNA methylation, methyl groups (see Fig. 1) are attached to the DNA backbone. In the case of histone modifications, methyl groups (or other small complexes made primarily of carbon and hydrogen) attach to the proteins that help coil the ~ 6 feet of DNA present in each cell into a microscopic size. Small noncoding RNAs, which I study in my research, regulate gene expression by preventing mRNA from being translated into protein. Very short pieces of RNA that are used in their RNA form rather than being turned into a protein (they are noncoding because they don’t code for a protein) bind to a mRNA transcript along with a protein complex called the RNA Induce Silencing Complex (RISC). Together the small RNA and the RISC keep only specific genes that have a sequence that is complementary to the to small RNA from being translated into a protein.

The human body has many parts, but the many parts make up one whole body. So it is with the body of Christ. (1 Corinthians 12:12 NLT)
These mechanisms are important for creating a phenotype because a single genotype contains instructions for multiple phenotypes. Within an individual being (human, water flea, or orchid), well-regulated gene expression is necessary for giving each part of the body its unique form and function through a process called differentiation. During differentiation stem cells, which have the potential to become any type of cell, become limited in their ability to express certain genes as regions of the DNA are methylated. The phenotype of a cell in the liver is very different from the phenotype of a kidney, muscle, or nerve cell even though they all contain the same genome. Unregulated gene expression causes chaos. In fact, cancers, diabetes, endometriosis, and other diseases are, at least partly, due to gene expression becoming dysregulated. Some birth defects can occur if genes fail to be turned on at the correct time.
In summary, I hope that you now have some understanding of the processes that turn a genotype into a phenotype, an appreciation for some of the mechanisms that regulate gene expression, and an awareness of the consequences of dysregulated gene expression. Stay tuned for the next installment when we will learn about animals that can have extremely different phenotypes contained within a genome, alternative developmental pathways. and how some of these same processes are important on a larger scale.
1. Deichmann, U. 2016. Epigenetics: The origins and evolution of a fashionable topic, Developmental Biology, 416: 249-254.
Dr. Julie A. Reynolds is a Research Scientist at The Ohio State University in the department of Evolution, Ecology, and Organismal Biology. She studies insect physiology and biochemistry with the goal of learning how animals adapt to extreme environments and survive changes in climate. In addition to writing for the Emerging Scholars Network, she is actively engages in discussions about science and faith as a Sinai and Synapses Fellow.

Leave a Reply