This week, I’m (virtually) attending the annual meeting of the Council of State and Territorial Epidemiologists (CSTE), one of the largest gatherings of folks doing the day-to-day work of public health in the United States. Yes, there is plenty of COVID-19 discussion, but that is far from the only topic. Each day there are hundreds of talks and posters about investigations, research, and applied science that will never make headlines. We are all probably more aware than we’d like to be about public health activities related to the pandemic. Still, I thought it might be good to share a glimpse of what else public health is up to all the time.
Bacterial resistance to antibiotics is a longstanding public health concern, especially resistance to potent antibiotics reserved for hardy strains that are not susceptible to first-choice therapies. So hospitals are regularly screening for resistance and reporting the results to public health, who follow up on cases of the more pernicious forms of resistance. On Monday, I attended a talk about one such investigation of a series of infections with bacteria resistant to carbapenems, one of those potent kinds of antibiotics. Thanks to advances in sequencing, it is becoming more common to sequence the entire genome of the bacteria isolated from such patients. This serves at least two purposes: providing insights into the mechanism of resistance–there are several ways to resist carbapenems–and identifying unique patterns of mutations that link bacteria across patients. Bacteria that differ by just ones or a few tens of mutations are likely to be very close relatives. If multiple patients have bacteria that closely related, those patients likely got infected from a common source or spread the bacteria one to another. The patients in this investigation were epidemiologically linked, meaning they were together in time and space in a way that would permit common exposure or transmission. But the genome analysis revealed two clusters of genomes, suggesting possibly two distinct sources. So which story is right?
The very question highlights one of the ways that public health is not just a scaled up version of medicine. Medically, the treatment for these patients is the same whether there was one source of infection or two. Maybe at some point it might become relevant if typical approaches were unsuccessful and an unusual therapeutic approach was required; then you might want to know how likely it would be that all patients would require the same therapy. But generally, physicians don’t need to know where the bacteria came from. For public health, however, one source or two is an important distinction. If there is an environmental source that needs to be cleaned or controlled, it is important to know if there is one cleaning project or two. If transmission is occurring, infection control measures need to be implemented at the relevant site or sites. knowing which story is correct, the epidemiological one or the genomic one, is relevant to public health.
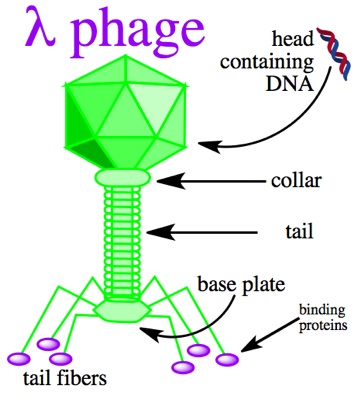
So, further investigation revealed that most of the differences between the two genome clusters traced to a single, small region of the genome. This is unlikely for mutations that have accumulated individually over time. When that one stretch of genome was removed from analysis, the two clusters looked like one, with just the small number of individual differences typical of bacteria recently derived from a single source. But you can’t just throw away inconvenient data; that small region needs to be reckoned with somehow. Fortunately, the region was small enough that its sequence could be uploaded to online tools that search sequence data from all kinds of organisms. That search revealed matches, but not from bacteria. The closest matches were all from a bacteriophage associated with this particular bacterial species. Bacteriophages are viruses that infect bacteria, and sometimes when they do some or all of their genome can get inserted into the bacterial genome. The investigators concluded such an insertion had occurred somewhere along the chain of transmission, possibly in the patient whose samples included bacteria with and without the bacteriophage genes. This provided a single story that makes sense of all the observations; the bacteria could all be related and still have acquired the observed changes in the short time allowed because what looked like a lot of small changes was really one big change.
Andy has worn many hats in his life. He knows this is a dreadfully clichéd notion, but since it is also literally true he uses it anyway. Among his current metaphorical hats: husband of one wife, father of two teenagers, reader of science fiction and science fact, enthusiast of contemporary symphonic music, and chief science officer. Previous metaphorical hats include: comp bio postdoc, molecular biology grad student, InterVarsity chapter president (that one came with a literal hat), music store clerk, house painter, and mosquito trapper. Among his more unique literal hats: British bobby, captain’s hats (of varying levels of authenticity) of several specific vessels, a deerstalker from 221B Baker St, and a railroad engineer’s cap. His monthly Science in Review is drawn from his weekly Science Corner posts — Wednesdays, 8am (Eastern) on the Emerging Scholars Network Blog. His book Faith across the Multiverse is available from Hendrickson.

Leave a Reply